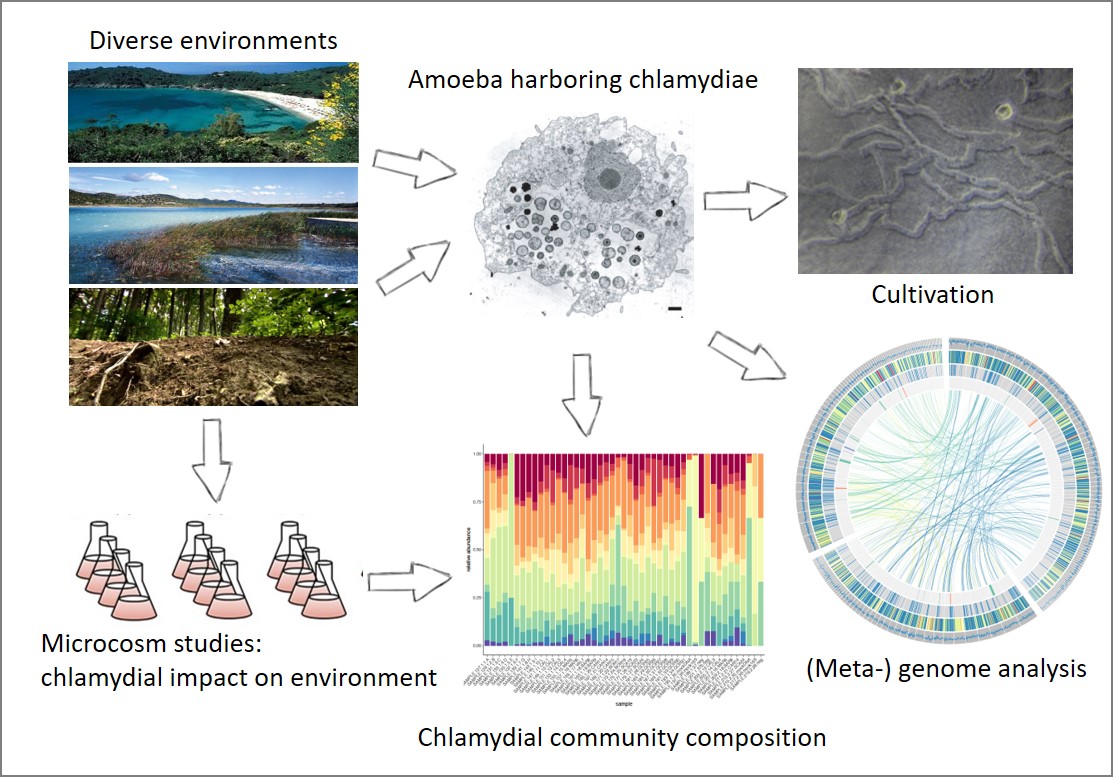
Characterization of natural chlamydial communities
Chlamydiae, such as Chlamydia trachomatis or C. pneumoniae, are well known bacterial pathogens of humans and animals. Only in the past 20 years closely related bacteria have been found in diverse environments where they thrive as symbionts in protists or as pathogens in a variety of animal hosts. The discovery and analysis of these environmental chlamydiae has provided new, exciting, and valuable insights into the basic biology and evolution of the whole phylum Chlamydiae. Yet, our knowledge about these microbes is mainly derived from in-depth analysis of only few representatives. Molecular surveys indicate a tremendous hidden diversity of chlamydiae in nature, and there is a remarkable gap concerning genomic information and isolates for the majority of environmental chlamydia lineages.
Aim: Characterization of natural chlamydial communities.
Based on the assumptions that (i) Chlamydiae are ubiquitous in nature, but their abundance and community composition differ between habitats and are affected by seasonal shifts and host dynamics, and (ii) The impact of chlamydiae on their protist hosts alters grazing behavior and thus contributes to shaping natural microbial communities and ecosystem functioning.
In addition, selected novel environmental chlamydiae will be investigated to provide a more comprehensive overview over variations of chlamydial biology and the evolution of the host-associated lifestyle.
Approach: Natural chlamydial communities from diverse environments will be investigated by combining a novel targeted amplicon sequencing assay, single cell and metagenomics, and isotope probing experiments with protist isolation approaches and microcosm experiments.
Relevance: The project will reveal the first comprehensive insights into chlamydial community structure and dynamics in nature. For the first time, the effect of chlamydiae on microbial food webs, community composition, and general microbial growth rate and mineralization will be determined. The recovery of novel chlamydiae and state-of-the-art single cell genomics will expand the knowledge about chlamydial biology, genome flexibility, and evolution.
Student: Angelika Schwarzhans
Faculty: Collingro (PI), Horn
Funding: FWF stand-alone project FunChlam