Genome-metabolome modeling and metabolome predictions in microbial symbioses
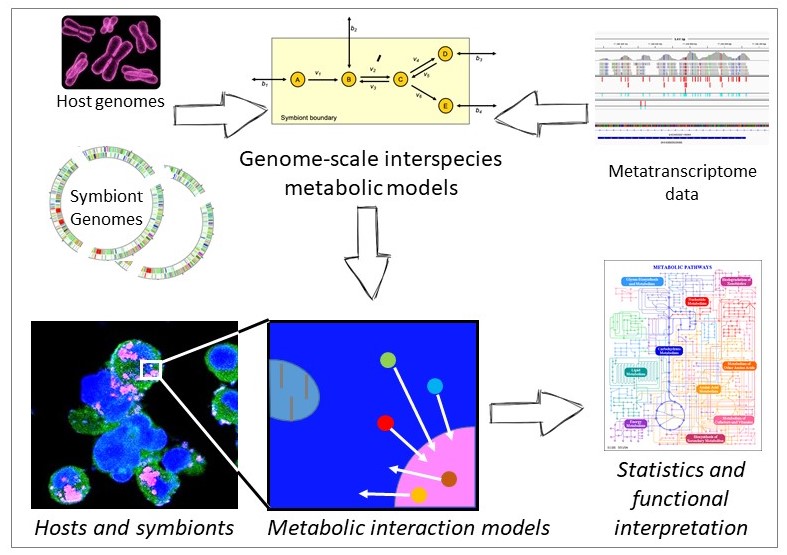
Metabolites are central for host-microbe and microbe-microbe interactions. Whole genome or metagenomic bin-based predictions of bacterial metabolism could not yet reasonably predict metabolites, which are central for host-microbe and microbe-microbe interactions.
Aims: We propose that lack of predicted proteins in incomplete metagenomic bins, to certain extent, could be compensated by computational gap filling algorithms. We further propose adding metatranscriptomic data to constrain possible solutions and allow to predict condition-dependent metabolomes. We hypothesize that genome-based predictions of metabolomes facilitate the linking of metabolites with bacterial species and/or hosts, which leads to function-level predictions that can be tested experimentally. We aim for the development of a metabolome prediction method based on a combination of automatic creation of metabolic models, flux-balance modeling and additional constraints due to metatranscriptomic data.
Approach: Metagenome sequencing, metatranscriptome sequencing, metabolomics, bioinformatics.
Relevance: Computational methods for multi*omics data analysis are urgently needed for a better interpretation of existing or upcoming data, not only in microbial ecology. In particular, we will go beyond statistical correlation analysis, seeking for a functional understanding of meta*omic data.
Student: Michael Predl
Faculty: Rattei (PI), Wagner, Woebken, Horn, Petersen
Funding: FWF doc.fund project MAINTAIN
Selected Publications:
Sczyrba, A., Hofmann, P., Belmann, P., Koslicki, D., Janssen, S., Dröge, J., Gregor, I., Majda, S., Fiedler, J., Dahms, E., Bremges, A., Fritz, A., Garrido-Oter, R., Jørgensen, T. S., Shapiro, N., Blood, P. D., Gurevich, A., Bai, Y., Turaev, D., DeMaere, M. Z., Chikhi, R., Nagarajan, N., Quince, C., Meyer, F., Balvočiūtė, M., Hansen, L. H., Sørensen, S. J., Chia, B. K. H., Denis, B., Froula, J. L., Wang, Z., Egan, R., Don Kang, D., Cook, J. J., Deltel, C., Beckstette, M., Lemaitre, C., Peterlongo, P., Rizk, G., Lavenier, D., Wu, Y.-W., Singer, S. W., Jain, C., Strous, M., Klingenberg, H., Meinicke, P., Barton, M. D., Lingner, T., Lin, H.-H., Liao, Y.-C., Silva, G. G. Z., Cuevas, D. A., Edwards, R. A., Saha, S., Piro, V. C., Renard, B. Y., Pop, M., Klenk, H.-P., Göker, M., Kyrpides, N. C., Woyke, T., Vorholt, J. A., Schulze-Lefert, P., Rubin, E. M., Darling, A. E., Rattei, T. & McHardy, A. C. Critical Assessment of Metagenome Interpretation-a benchmark of metagenomics software. Nat. Methods 14, 1063–1071 (2017).
Sato, Y., Ling, E. Y. S., Turaev, D., Laffy, P., Weynberg, K. D., Rattei, T., Willis, B. L. & Bourne, D. G. Unraveling the microbial processes of black band disease in corals through integrated genomics. Sci. Rep. 7, 40455 (2017).
Güllert, S., Fischer, M. A., Turaev, D., Noebauer, B., Ilmberger, N., Wemheuer, B., Alawi, M., Rattei, T., Daniel, R., Schmitz, R. A., Grundhoff, A. & Streit, W. R. Deep metagenome and metatranscriptome analyses of microbial communities affiliated with an industrial biogas fermenter, a cow rumen, and elephant feces reveal major differences in carbohydrate hydrolysis strategies. Biotechnol. Biofuels 9, 121 (2016).
Turaev, D. & Rattei, T. High definition for systems biology of microbial communities: metagenomics gets genome-centric and strain-resolved. Curr. Opin. Biotechnol. 39, 174–181 (2016).
Feldbauer, R., Schulz, F., Horn, M. & Rattei, T. Prediction of microbial phenotypes based on comparative genomics. BMC Bioinformatics 16 Suppl 14, S1 (2015).